Towards supervised biomedical semantic similarity
- Conference/Journal: SeWeBMeDA 2022
- Year: 2022
- Full paper URL: assets/papers/sss_SeWeBMEDA.pdf
Abstract
Ontology-based semantic similarity between entities in knowledge graphs is essential for several bioinformatics applications, including the prediction of protein-protein interactions and the discovery of associations between diseases and genes. Knowledge graphs typically describe entities according to different aspects modeled in ontologies, but both classical and graph embeddings-based semantic similarity measures consider the graph as a whole. This can be a limitation since different use cases may require different similarity perspectives and ultimately depend on expert knowledge for manual fine-tuning.
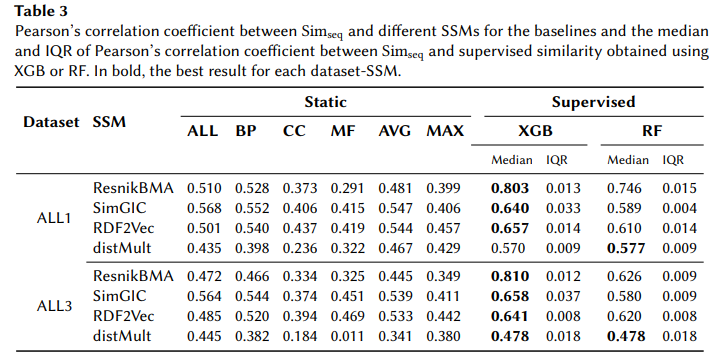
We present a new approach that uses supervised machine learning to tailor aspect-oriented semantic similarity measures to fit a particular view on biological similarity. This results in a supervised semantic similarity that is independent of the downstream application. We implement and evaluate it using different combinations of representative semantic similarity measures and machine learning methods with three biological similarity views: protein function family similarity, protein sequence similarity and phenotype-based gene similarity.
The results demonstrate that our approach outperforms non-supervised methods, producing semantic similarity models that fit different biological perspectives significantly better than the commonly used manual combinations of semantic aspects.