Multi-dataset and Transfer Learning Using Gene Expression Knowledge Graphs
- Conference/Journal: Extended Semantic Web Conference (ESWC)
- Year: 2025
- Full paper URL: https://link.springer.com/chapter/10.1007/978-3-031-94575-5_14
Abstract
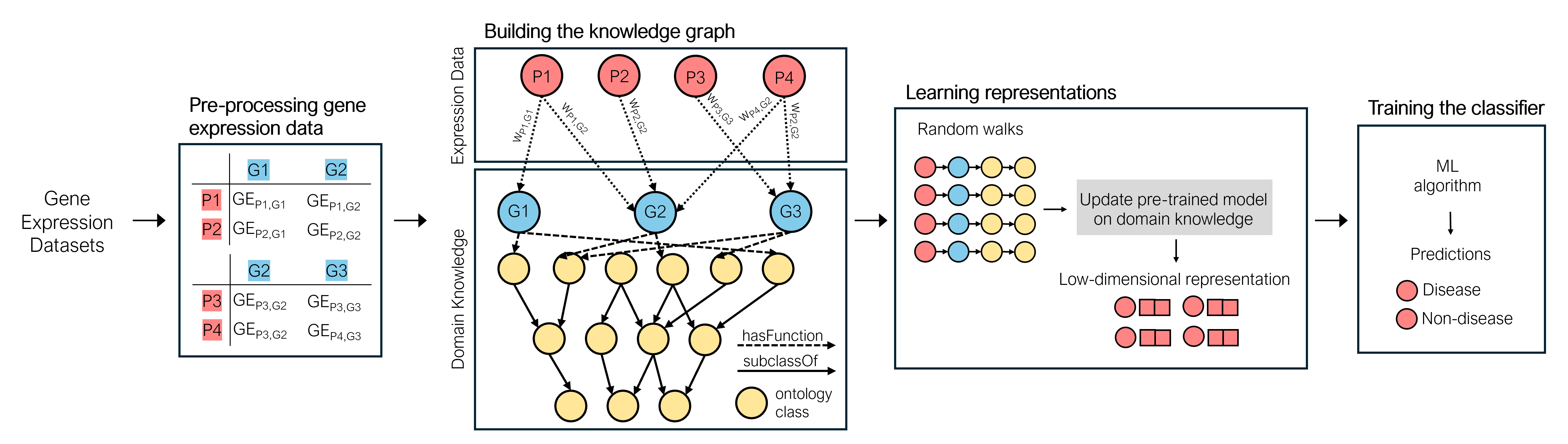
Gene expression datasets offer insights into gene regulation mechanisms, biochemical pathways, and cellular functions. Additionally, comparing gene expression profiles between disease and control patients can deepen the understanding of disease pathology. Therefore, machine learning has been used to process gene expression data, with patient diagnosis emerging as one of the most popular applications. Although gene expression data can provide valuable insights, challenges arise because the number of patients in expression datasets is usually limited, and the data from different datasets with different gene expressions cannot be easily combined.
This work proposes a novel methodology to address these challenges by integrating multiple gene expression datasets and domain-specific knowledge using knowledge graphs, a unique tool for biomedical data integration. Then, vector representations are produced using knowledge graph embedding techniques, which are used as inputs for a graph neural network and a multi-layer perceptron. We evaluate the efficacy of our methodology in three settings: single-dataset learning, multi-dataset learning, and transfer learning. The experimental results show that combining gene expression datasets and domain-specific knowledge improves patient diagnosis in all three settings.