Gene Expression Knowledge Graph for Patient Representation and Diabetes Prediction
- Conference/Journal: Journal of Biomedical Semantics
- Year: 2025
- Full paper URL: https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-025-00325-6
Abstract
Diabetes is a worldwide health issue affecting millions of people. Machine learning methods have shown promising results in improving diabetes prediction, particularly through the analysis of gene expression data. While gene expression data can provide valuable insights, challenges arise from the fact that the number of patients in expression datasets is usually limited, and the data from different datasets with different gene expressions cannot be easily combined
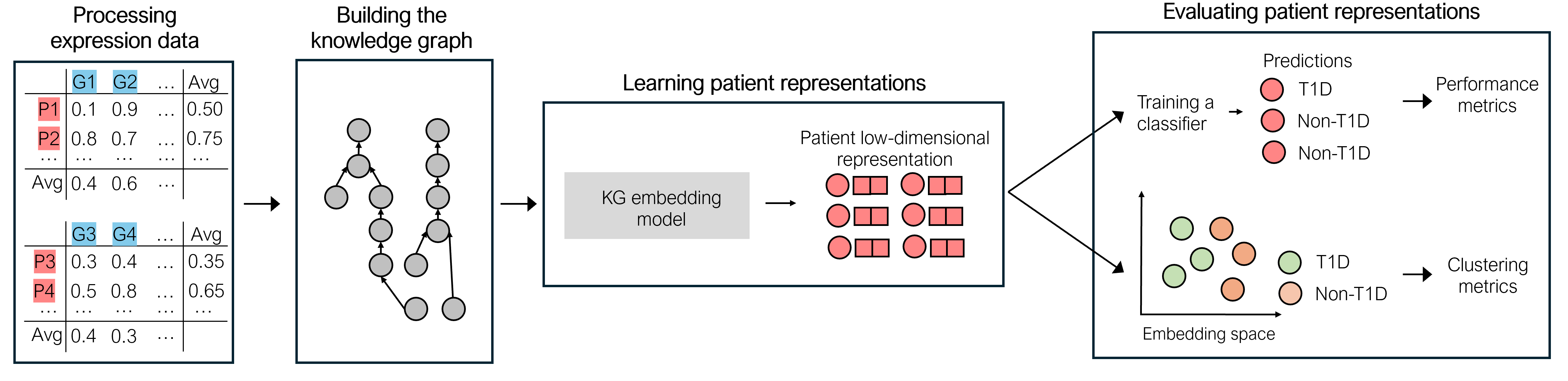
This work proposes a novel approach to address these challenges by integrating multiple gene expression datasets and domain-specific knowledge using knowledge graphs, a unique tool for biomedical data integration, and to learn uniform patient representations for subjects contained in different incompatible datasets. Different strategies and KG embedding methods are explored to generate vector representations, serving as inputs for a classifier. Extensive experiments demonstrate the efficacy of our approach, revealing weighted F1-score improvements in diabetes prediction up to 13\% when integrating multiple gene expression datasets and domain-specific knowledge about protein functions and interactions.
This paper builds upon our prior work presented at the 7th Workshop on Semantic Web Solutions for Large-scale Biomedical Data Analytics, co-located with the Extended Semantic Web Conference 2024. This version contains various extensions, including the comparison of different embedding variants, and a more thorough experimental evaluation of the cross-dataset class separation capabilities of the different patient representation approaches.