Integrating heterogeneous gene expression data through knowledge graphs for improving diabetes prediction
- Conference/Journal: SeWeBMeDA 2024
- Year: 2024
- Full paper URL: assets/papers/diabetes_SeWeBMEDA.pdf
Abstract
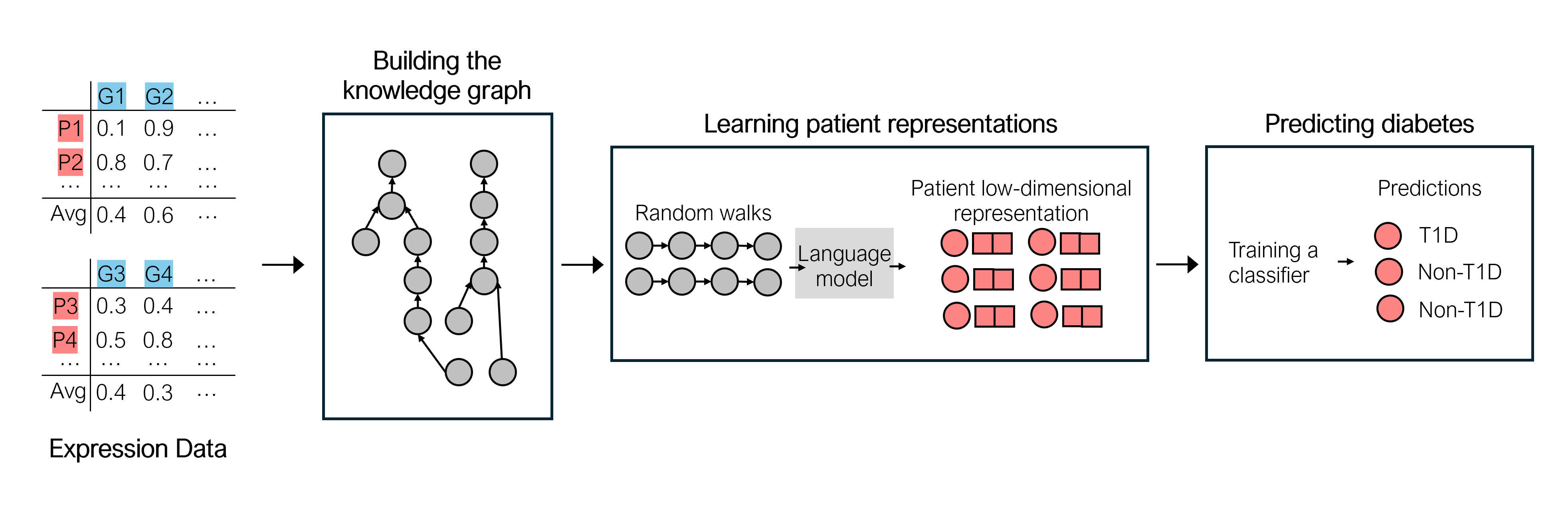
Diabetes is a worldwide health issue affecting millions of people. Machine learning methods have shown promising results in improving diabetes prediction, particularly through the analysis of diverse data types, namely gene expression data. While gene expression data can provide valuable insights, challenges arise from the fact that the sample sizes in expression datasets are usually limited, and the data from different datasets with different gene expressions cannot be easily combined.
This work proposes a novel approach to address these challenges by integrating multiple gene expression datasets and domain-specific knowledge using knowledge graphs, a unique tool for biomedical data integration. KG embedding methods are then employed to generate vector representations, serving as inputs for a classifier. Experiments demonstrated the efficacy of our approach, revealing improvements in diabetes prediction when integrating multiple gene expression datasets and domain-specific knowledge about protein functions and interactions.